INTRODUCTION
Grasses (Poaceae) are the fifth largest family of flowering plants in the world, with approximately 11,500 species and about 768 genera (Soreng et al., 2017). The grass family is economically, ecologically, and evolutionarily one of the most successful species-rich groups, due to which it represents as a model family for the study of speciose taxa (Trevor, 2018). A wide range of environmental stress such as drought, high salt, and temperature affects plants all over the world. Due to these environmental changes and stress exerted on the crops, the growth and yield are very much affected. Worldwide, there is about 50% reduction of average yield, and 10% is due to the environmental stresses, particularly drought and salinity (Bray et al., 2000). Response to abiotic stresses is a very complex phenomenon as various stages of plant development can be affected by a particular stress and often several stresses simultaneously affect the plant (Chinnusamy et al., 2004). Dehydration-responsive element-binding protein (DREB) is a type of plant-specific transcription factor which specifically binds to DRE/CRT elements in the response to abiotic stresses such as drought and low temperature and their function was identified in Arabidopsis (Gilmour et al., 1998; Liu et al., 1998) at first. DREB1 and DREB2 genes have specific roles in Arabidopsis like it can improve the tolerance to drought, and it can increase the survival rate under cold stress, respectively.
METHODOLOGY
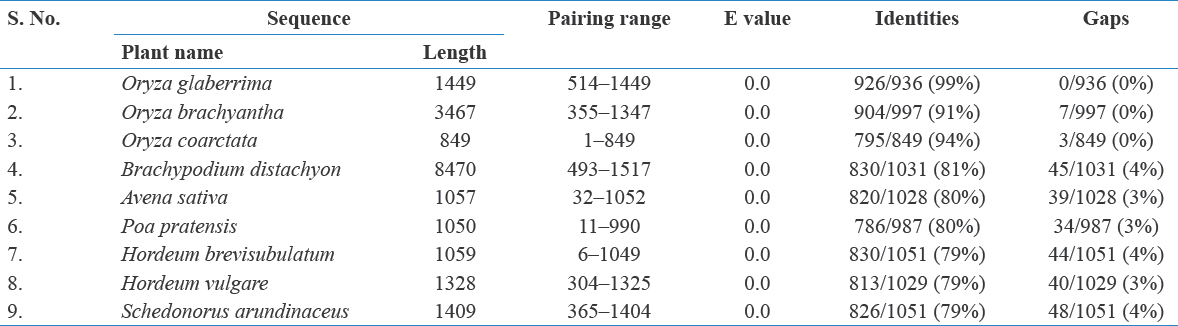
BLAST was done with Oryza sativa (Acc. no: AY064403.1) nucleotide sequence for DREB gene. The hits list was studied and nine plants (E-value – 0) belonging to the family Poaceae were selected. Their nucleotide sequences was retrieved from NCBI in FASTA format, and further analysis was performed using MEGA X software for the 10 nucleotide sequences (O. sativa, Oryza glaberrima, Oryza brachyantha, Oryza coarctata, Brachypodium distachyon, Avena sativa, Poa pratensis, Hordeum vulgare, Hordeum brevisbulatum, and Schedonorus arundinaceus). To study the positions placed according to taxonomic details among the selected plants, NCBI taxonomic browser was used. Alignment and phylogenetic study was conducted using software Mega X (Tramura et al., 2013). Alignment is done using Clustal W method. Phylogenetic tree was obtained using maximum likelihood method.
RESULTS AND DISCUSSION
The present study deals with the evolutionary aspects of few selected plants with respect to DREB gene through simple phylogenetic analysis. Nucleotide sequence for DREB gene in O. sativa was selected and BLAST was run. Sequence selection from the BLAST hits for further analysis is done based on e value (0) and from plants belonging to Poaceae. The details of pairwise alignment with the query sequence are given in Table 1. FASTA format of selected nucleotide sequences was downloaded for further analysis using MEGA X.
Table 1: Results of pairwise alignment with Oryza sativa
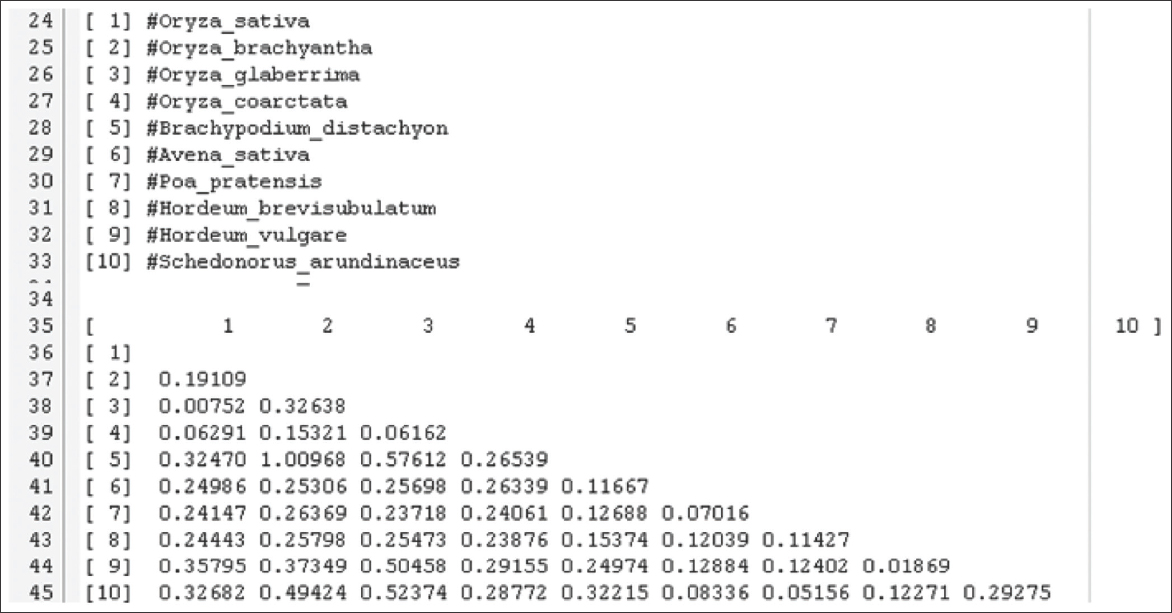
Multiple sequence alignment using Clustal W was done using Molecular Evolution Genetic Analyzer (MEGA X) software. Pairwise distance analysis was conducted using the maximum composite likelihood model (Tamura et al., 2004) [Figure 1] which shows that Oryza glaberrima and O. sativa are close, while H. vulgare and O. sativa are found to be far distantly placed.
Figure 1: Pairwise distance data of evolutionary divergence
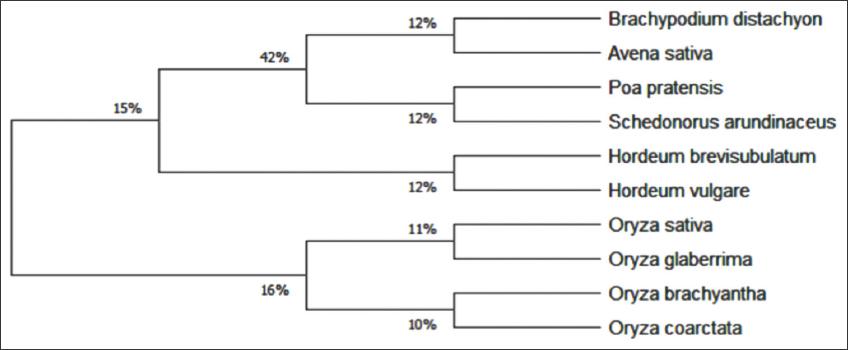
The evolutionary history was inferred using the minimum evolution method (Rzhetsky and Nei, 1992). The optimal tree with the sum of branch length = 0.92997563 is shown in Figure 2. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. The ME tree was searched using the close-neighbor-interchange algorithm (Nei and Kumar, 2000) at a search level of 1. The neighbor-joining algorithm (Saitou and Nei, 1987) was used to generate the initial tree. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018) and the result shows the same tree division of two main branches of plants belonging to Pooideae and Oryzoideae subfamily. Taxonomic browser results also indicated that all 10 plants belonged to BOP clade under two subfamilies Pooideae (B. distachyon, A. sativa, P. pratensis, H. vulgare, H. brevisbulatum, and S. arundinaceous) and Oryzoideae (O. sativa, O. glaberrima, O. brachyantha, and O. coarctata).
Figure 2: Evolutionary relationships of taxa taken for the study
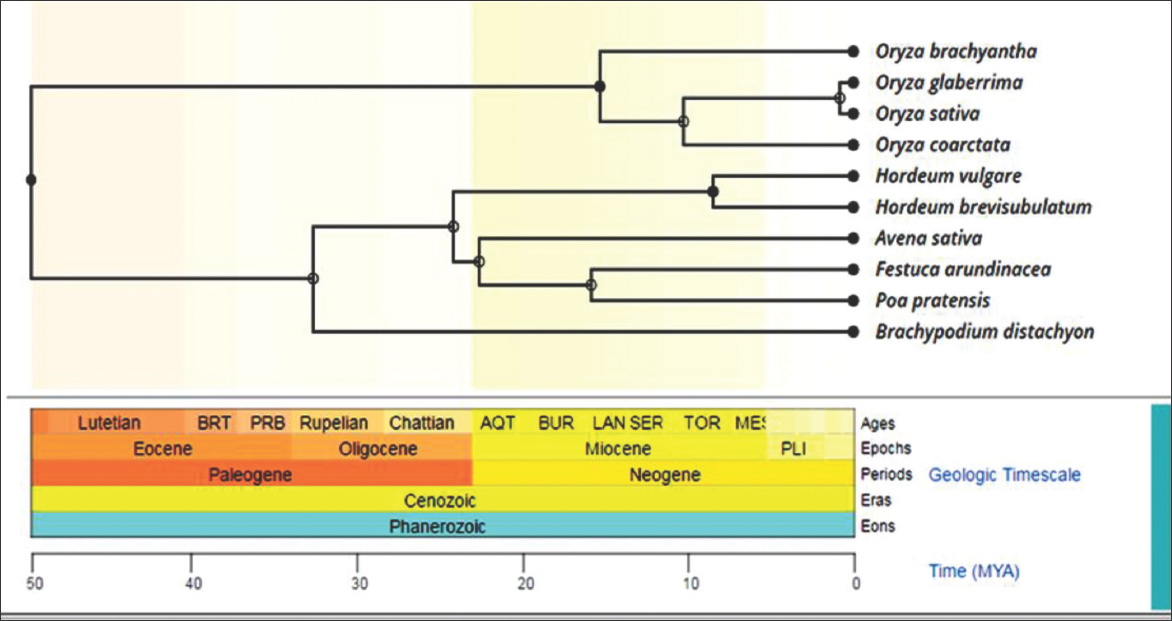
Timeline or time tree is performed for the 10 selected plants [Figure 3] using MEGA X software. It shows that genus Oryza has evolved during Lutein ages, while the species of Oryza evolved during Miocene epochs. O. glaberrima and O. sativa were found to get evolved PLI epochs. O. sativa and O. glaberrima are Asian and African rice, respectively, and according to Ndjiondjop et al. (2018), both have a narrow genetic diversity.
Figure 3: Time line tree for the 10 plants selected for the study
CONCLUSION
The plants selected for this study belong to BOP clade and subfamilies Pooideae and Oryzoideae. Taxonomic browser tree and the tree generated through MEGA X software using DREB gene showed that the two subfamilies were grouped together. These results show that DREB gene is a homologous gene among the selected plants taken for this study.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
The authors declare that there is no conflict of interest in this work.