INTRODUCTION
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) came from the Chinese seafood in the town of Wuhan, has led to the rapid transmission to 210 countries worldwide (Din and Boppana, 2020). All over the world, scientists are struggling to understand this new and rapidly spreading virus that causes SARS-CoV-2 with the aim of developing effective involvements for controlling and preventing CoV, which cause various harmful diseases (Zhong et al., 2003). CoVs belong to the order Nidovirales, are mainly enveloped viruses with the largest single-stranded RNA and genome size of 26–32 kb in length (Zaki et al., 2012). The SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) also belong to beta CoVs that are zoonotic in origin and linked to potentially fatal illness during the outbreaks in 2003 and 2012, respectively. Due to the rapid increase of SARS-CoV-2 infections and affected countries, efforts for developing an effective SARS-CoV-2 vaccine are crucially required. CoVs encode multiple structural and non-structural proteins. The structural proteins include the spike (S) protein, the envelope (E) protein, the membrane (M) protein, and the nucleocapsid (N) protein. With SARS-CoV-2 being discovered recently, the information about immunogenic epitopes eliciting antibody or T-cell responses is relatively less. Studies suggest that SARS- CoV-2 is similar to SARS-CoV-1 which is based on phylogenetic analysis, complete genome, binding of the human cell receptor and similarity in the mechanism of cell entry (Letko et al., 2020 and Zhou et al., 2020) Concerning the similitude between the two infections, the past examinations additionally gave an intelligent defensive resistant reaction against SARS-CoV-1 that can conceivably energize antibody plan for SARS-CoV-2. Exploration additionally has indicated that the immunizer reactions created against the S protein, the most uncovered protein of SARS-CoV, can be protective against contamination in creature models (Yang et al., 2004). Besides, various investigations have demonstrated that antibodies are delivered against the SARS-CoV protein N, an exceptionally immunogenic protein and generally communicated, during contamination (Ying et al., 2003). Besides, examines have shown that SARS-CoV-2 inspires a strong B-cell reaction and this safe humoral reaction causes the freedom of SARS-CoV-2 [Figure 1]. Lymphocytes, moreover, assume a principal part in viral contaminations. For instance, CD4 immune system microorganisms give B-cell help to counteracting agent creation, which prompts enduring security. Henceforth, the improvement of antibodies against SARS-CoV-2 that can animate B and lymphocytes is profoundly valuable.
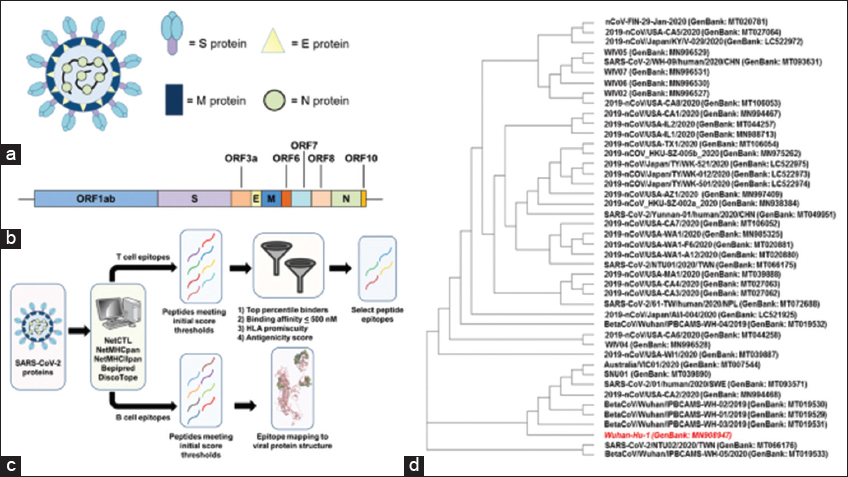
Figure 1: (a) Diagram of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virion structure with the major structural proteins (S, M, N, and E) highlighted. (b) Cartoon representation of the SARS-CoV-2 genome with the 10 major protein-coding regions annotated. The box diagrams are proportional to the protein size. (c) Diagram of peptide identification workflow illustrating the algorithms used 36,44,45,46,47,49,50,51,58,60 and filtering criterion applied to refine peptide selection. (d) Cladogram illustrating the genetic relationship of SARS-CoV-2 isolates. The original viral isolate and consensus sequence (Wuhan-Hu-1) are highlighted in red
From one viewpoint, as peptide-based immunizations do not require in vitro culture of pathogenic infection, they are protected, and their selectivity permits precise enactment of resistant reactions through conventional antibodies which are expensive, allergenic, tedious, and furthermore dangerous because they need in vitro culture (Abdelmageed et al., 2020). Then again, immunizer subordinate improvement (ADE) of viral passage, immunization advancement, and antibody-based drug treatment has been among significant worries for disease transmission specialists in managing numerous infections. ADE of infection disease is a wonder in which explicit infection antibodies improve the passage of infection and, at times, the replication of infection into insusceptible cells through collaboration with Fc and supplement receptors. ADE has been noticed in COVID for quite a long time, and because of the ongoing advancement toward understanding the receptor acknowledgment and film combination systems of COVID spikes, COVID speaks to a magnificent model framework for researching ADE of viral diseases. Concerning the writing on different SARS-CoV-1 and MERS-CoV immunization endeavors which have bombed because of ADE in creature models, it is sensible to speculate a comparable ADE hazard for SARS-CoV-2 antibody endeavors except if they explicitly target areas that will obstruct infection insusceptible cell combination.
Further, a few examinations have exhibited that killing MAbs focusing on different pieces of viral spikes would be less inclined to intervene ADE on the off chance that they do not trigger the conformational changes of the spikes. Consequently, to lessen the probability of ADE, spike-based subunit antibodies lacking the receptor-restricting area (RBD) can be intended to forestall viral contaminations.
Through this survey, we have endeavored to gather the data that are thoroughly coordinating and contain no change in the accessible SARS-CoV-2 arrangements. These epitopes can possibly instigate a viable reaction against SARS-CoV-2. The point of this thorough audit (SR) is to assist scientists with creating antibodies by gathering information for the control and anticipation of SARS-CoV-2 and the resistant and bioinformatic ID of lymphocyte and B-cell epitopes.
METHODS
The present review was conducted over the research studies published in the protein structure of SARS-CoV-2. In developing the systematic on immune and bioinformatics identification of T-cell and B-cell epitopes review (SR) protocol, preferred reporting elements for the SR statements, meta-analyses (PRISMA), and guidelines from the Cochrane Reviewer’s Handbook were used (http://www.prisma-statement.org/Extensions/InDevelopment.aspx) (Mohamoud et al., 2013, Noorimotlagh et al., 2020), (Noorimotlagh et al.), (Noorimotlagh et al., 2018).
Information Sources and Search Strategy
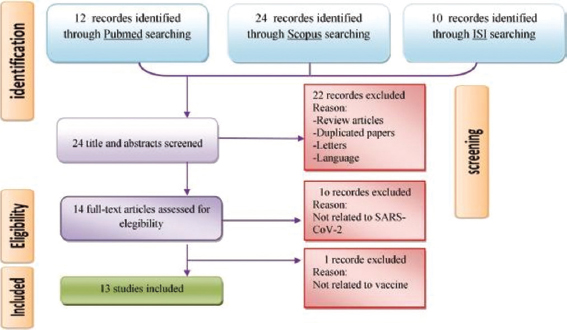
We played out a precise bibliographic pursuit during 2019–2020. The keep going hunt was led on April 24, 2020. Organization for Logical Data (ISI) Web Science, Scopus, MEDLINE, and Google Researcher information bases were utilized to look through utilizing cross-section (Clinical Subject Headings), free content words, and all conceivable mix. The accompanying legitimate catchphrases were utilized: (“nCov” OR “Novel COVID” OR “2019 Novel COVID” OR “CoVs” OR “2019-nCoV” OR “Extreme Intense Respiratory Condition COVID 2” OR “SARS-CoV-2”) AND (“B-cell” OR “Lymphocyte” OR “Epitope” OR “Peptide” OR “Antibody,” as outlined in Figure 2. Consideration/avoidance measures for the included examinations articles were efficiently checked on, and distributions were chosen dependent on the accompanying models: Articles should have been (a) written in English, (b) unique papers, (c) electronically accessible (on the web), and (d) vaccine-centered exploration for SARS-CoV-2 and distinguishing proof bioinformatics of lymphocyte and B-cell epitopes. The book audit, book parts, rules, survey articles, copy articles, different dialects (French, German, Italian, and Spanish), letters to editors, short interchanges, oral introduction, gathering records, and remarks were considered as exclusion standards in this deliberate audit. By and large, we inspected articles that presented epitopes that were distinguished by B and lymphocytes and were separated from viral antigenic proteins, for example, S, M, N, and E. Likewise, these chose epitopes should have been investigated regarding antigenicity, allergenicity, and physiological properties and were introduced as powerful epitopes in inciting resistant reactions.
Figure 2: Summary of a standard four-step protocol for literature review
Data Extraction
Two reviewers (SM and MA) independently investigated the titles, abstracts, and full texts from each database. Considering each selected study, items such as first author, country, year of publication, type of protein, B-cell/T-cell epitope, antigenicity, HLA Class I, HLA Class II, score, and results were extracted.
RESULTS
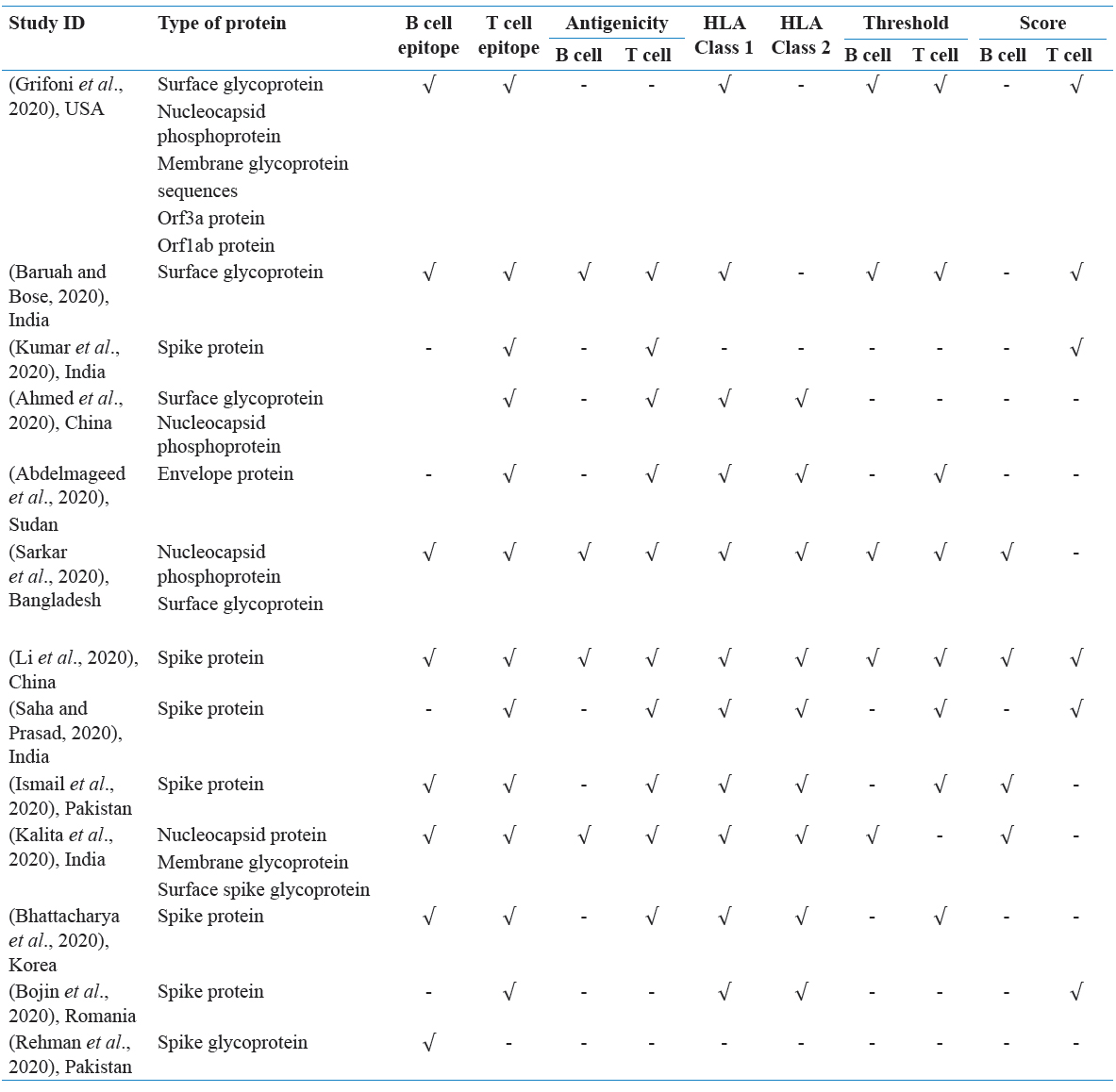
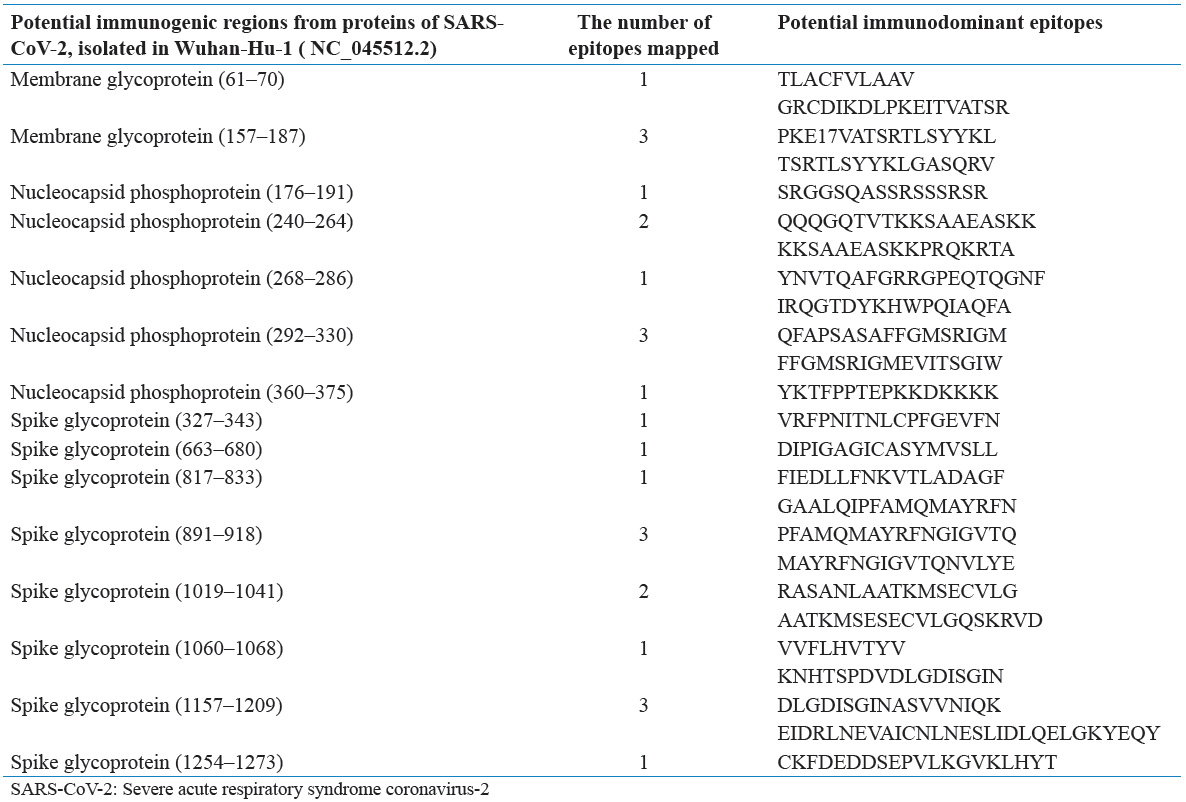
In this SR, 166 articles were recognized through the underlying quests. In light of title and dynamic, 22 articles were barred on the grounds that they were either copy, audits, letters, or written in different dialects. Eleven full-text papers were likewise avoided in light of the fact that they were irrelevant to SARS-CoV-2 and antibody. At long last, a sum of 13 remarkable papers was incorporated with the end goal of this exploration. Figure 1 represents the stream graph of the choice of “bit by bit” examines and the number of studies recognized for this audit. Table 1 sums up a nitty-gritty portrayal of the 13 articles inspected. Next, all the chose articles were separated by the attributes of B and lymphocyte epitopes distinguished in SARS-CoV-2 (Tables 2 and 3).
Table 1: The point mutations found within the potential epitopes of American SARS-CoV-2 isolates
Table 2: Summary of the main detailed of the included studies
Table 3: Potential immunodominant regions of SARS-CoV-2 and the mapped epitopes in those regions
DISCUSSION
The episode of the new COVID (SARS-CoV-2) in China and its ensuing pandemic raised an extraordinary arrangement of worldwide concerns. Binding this viral contamination in human networks requires sufficient data on its transmission courses, seriousness, and confusions, just as admittance to proper therapeutics and immunizations (Kumar et al., 2020). Endeavors to build up a successful antibody against the SARS-CoV-2 mean to restrict and afterward kill the disease from human social orders. Nonetheless, creating a successful SARS-CoV-2 immunization is risky because of the absence of adequate data about its organic qualities and insusceptible reactions against it (Ahmed et al., 2020). In this way, it appears to be that the immunoinformatics-based methodologies are needed to research the immunogenic epitopes and immunization configuration utilizing information from proteins sequencing of the Co-Vs. The SARS-CoV-2 is a RNA infection with four basic proteins; S (Spike), E (Envelope), N (Nucleocapsid), and M (Film). These basic proteins assume significant functions in the passage of the infection into having cells and ensuing compartmentation of viral particles. Safe reactions, including immunizer creation against the auxiliary proteins, are basic to restrict the spread of the contamination. In this manner, endeavors have been devoted to plan and deliver peptide antibodies focusing on immunogenic epitopes on the basic viral proteins to push resistant cells to rapidly distinguish these significant viral epitopes (Kumar et al., 2020, Sun, 2020). T and B lymphocytes are two significant kinds of invulnerable cells in the body (Baruah and Bose, 2020, Sarkar et al., 2020). Endless supply of an antigen into the body, it is coordinated to aid immune system microorganisms through MHC-II atoms communicated by antigen-introducing cells (APCs). At the same time, partner immune system microorganisms express the CD4 surface marker, another subset of lymphocytes, known as cytotoxic white blood cells, expresses CD8. Antigens are introduced to these cells, however, MHC-I particles. Then again, enacted assistant white blood cells instigate B lymphocytes which at last discharge a lot of killing antibodies.
Subsequent counteracting agent subordinate improvement (ADE) of viral passage has been identified for some infections. It was uncovered that antibodies chose one serotype of infections as well as sub-kill extra, head ADE of the last infections (de Alwis et al., 2020). The first system for ADE incorporates a killing immune response official to the infection surface structure protein of COVID such as a viral receptor, initiating a conformational alteration of the spike, and encouraging viral section into IgG-Fc-receptor-communicating cells through perceived viral-receptor-dependent pathways. Despite the consecutive connection between the timings of serious SARS and immune response extension, other potential explanations for the recognized pathology have not been deductively proved. Previous counteracting agent development could be dictated by prepping impact from past human CoV contaminations with no impact on pathogenesis, paying little heed to the time-sensitive incident with other extreme sicknesses. In addition, the energy of immunizer reaction is additionally known to be impacted by viral burden and intrinsic safe responses (Wan et al., 2020). The aide cells likewise enact cytotoxic immune system microorganisms, which straightforwardly murder contaminated cells (Sarkar et al., 2020). Thusly, it is critical to recognize the SARS-CoV-2 infection epitopes that prompt productive T-assistant, T-cytotoxic, and B-cells to create immunizations against the infection. SARS-CoV-2 nearest relative among human COVID is SARS-CoV, with 79% hereditary similarity. There is 72% similitude in the corrosive amino succession of the RBDs of SARS-CoV and SARS-CoV-2, with exceptionally comparable 3D structures. Homology displaying and biophysical properties assign that the SARS-CoV-2 RBD space ties to ACE2 with 10–20 crease higher partiality than that of SARS-CoV (Tay et al., 2020).
Many bioinformatics studies have been conducted in recent months to determine epitopes that are detected by T and B cells. We have introduced a portion of these investigations in Tables 1-3. In these examinations, the proteins of CoV infection that has a basic function in the destructiveness of the infection were chosen, and afterward, its antigenic properties were inspected. After the antigenic investigation of these proteins, objective proteins, for example, S or N proteins, were chosen for conclusive examination. The invulnerable epitope information base or IEDB that contains a tremendous assortment of exploratory information on immune system microorganism epitopes was utilized for the expectation of epitope of partner white blood cell dependent on their percentile rank and the middle inhibitory focus (IC50) values. Likewise, NetCTL 1.2 and NetMHC skillet EL 4.0 workers were utilized for the expectation of CTL epitope (Sarkar et al., 2020). Epitopes recognized by lymphocytes are short peptide sections that must be assessed for their capacities to tie MHC-I and MHC-II atoms. The epitopes chose for creating immunizations ought to likewise have a high fondness for MHC-I and II atoms and present the most noteworthy total populace inclusion (Ahmed et al., 2020). Direct epitopes distinguished by B-cells are commonly more than those recognized by lymphocytes and can be pieces of auxiliary proteins. Such epitopes, if being immunogenic, can initiate more powerful humoral reactions against viral diseases. For the forecast of straight B-cell epitopes in SARS-CoV-2 surface glycoprotein, nucleocapsid phosphoprotein, and layer glycoprotein, these investigations utilized BepiPred 2.0 and ABCPred workers (Basu et al., 2020).
The designed vaccine candidate should characterize excessive antigenic property to be capable of stimulating fine immune responses toward viral infections except for the want for any adjuvant and additionally need to no longer set off an allergic response to the body. Hence, the epitopes chosen in the preceding step in this research have been examined in phrases of antigenicity (using the VaxiJen server), allergenicity (using the AllerTOP server), and physicochemical parameters (using ProtParam server). Finally, epitopes with excessive antigenicity and except allergenic or poisonous results had been delivered as workable goals for growing peptide vaccines toward SARS-CoV-2 virus. Advanced epitope maps (T-cell and B-cell tests) are required to determine on the achievable of diagnosed epitopes to set off a fine resistant response in opposition to SARS-CoV-2. This would provide help to assist refine the specified set of the epitope, based totally on the immunogenicity inspected; a crucial idea for the immunogenic plan. Overall, as the uncommon set of SARS-CoV epitopes is indistinctly exceptional from SARS-CoV-2, they exhibit treasured possible candidacies for directing exploratory endeavors in the direction of developing immunizations toward SARS-CoV-2. More, for the most part, our deliberate help highlights the practicable significance of previous scientific and exploratory concerns for SARS-CoV and is used in conjunction with developing statistics for SARS-CoV-2 in the search for compelling immunizations to fight the COVID-19 outbreak. Characteristically, all vaccines had been created the usage of stay or weakened microorganisms or components of them.
However, the use of complete organisms, their aspects, or the organic method for vaccine manufacturing have numerous weaknesses. The presence of immunologically pushed aside organic elements or organic impurities in such vaccines may reason for primary complications. All the negative facets of typical vaccines may be incredulous by way of the improvement of completely artificial peptide-based vaccines. However, as soon as solely minimal antigenic epitopes are utilized for immunization, the immune responses are poor. The use of an adjuvant can overcome this hindrance; however, it may additionally amplify new malfunctions (Skwarczynski and Toth, 2020).
CONCLUSIONS
B and T cells are two important arms of the immune system against pathogens, including viruses. Activation of these cells plays an important role in the body’s defenses. These cells are activated in both natural active (pathogen entry) and artificial active (vaccine) forms. The vaccine is now being used to treat many infections. In the present study, we reviewed epitopes of structural proteins identified as effective and recognizable targets by immune (B and T) cells for SARS-CoV-2 virus. This can help early stages of vaccine development attempts to produce more effective peptide vaccines.
ACKNOWLEDGMENT
I express my gratitude to everyone who supported me and to Dr. Ramneet Kaur for the keen interest she took in me. She explained out solutions to the problems that had a straining effect. Without her help, it was a matter of acute impossibility to propel in this endeavor. It is a matter of profound privilege and pleasure to pay my sincere and heartfelt thanks to her. I am also very grateful to who edited the manuscript.
REFERENCES
1. Abdelmageed, M.I., Abdelmoneim, A.H., Mustafa, M.I., Elfadol, N.M., Murshed, N.S., Shantier, S.W. and Makhawi, A.M. 2020. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV:An immunoinformatics approach.
2. Ahmed, S.F., Quadeer, A.A. and McKay, M.R. 2020. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies.
3. Basu, A., Sarkar, A. and Maulik, U. 2002. Strategies for vaccine design for corona virus using Immunoinformatics techniques.
4. de Alwis, R., Chen, S., Gan, E.S. and Ooi, E.E. 2002. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development.
5. Din, M.A.U. and Boppana, L.K.T. 2020. An update on the 2019-nCoV outbreak.
6. Kumar, S., Maurya, V.K., Prasad, A.K., Bhatt, M.L.B. and Saxena, S.K. 2020. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV).
7. Letko, M., Marzi, A. and Munster, V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses.
8. Mohamoud, Y.A., Mumtaz, G.R., Riome, S., Miller, D. and Abu-Raddad, L.J. 2013. The epidemiology of hepatitis C virus in Egypt:A systematic review and data synthesis.
9. Noorimotlagh, Z., Mirzaee, S.A., Ahmadi, M., Jaafarzadeh, N. and Rahim, F. 2018. The possible DNA damage induced by environmental organic compounds:The case of Nonylphenol.
10. Noorimotlagh, Z., Mirzaee, S.A., Martinez, S.S., Rachoń, D., Hoseinzadeh, M. and Jaafarzadeh, N. 2020. Environmental exposure to nonylphenol and cancer progression Risk-A systematic review.
11. Noorimotlagh, Z., Zhang, Y., Qiao, W., Zhang, J and Qi, Z. 2002. Baricitinib, a drug with potential effect to prevent SARS-COV-2 from entering target cells and control cytokine storm induced by COVID-19.
12. Sarkar, B., Ullah, M.A., Johora, F.T., Taniya, M.A. and Araf, Y. 2020. The essential facts of Wuhan novel coronavirus outbreak in China and epitope-based vaccine designing against 2019-nCoV.
13. Tay, M.Z., Poh, C.M., Renia, L., Macary, P.A. and Ng, L.F. 2020. The trinity of COVID-19:Immunity, inflammation and intervention.
14. Wan, Y., Shang, J., Sun, S., Tai, W., Chen, J., Geng, Q., He, L., Chen, Y., Wu, J. and Shi, Z. 2020. Molecular mechanism for antibody-dependent enhancement of coronavirus entry.
15. Yang, Z., Kong, W., Huang, Y., Roberts, A., Murphy, B.R., Subbarao, K. and Nabel, G.J. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice.
16. Ying, L.I.N., Xu, S., Yang, R.F., Li, Y.X., Ji, Y.Y., He, Y.Y., De Shi, M., Wei, L.U., Shi, T.L. and Jin, W. 2003. Identification of an epitope of SARS-coronavirus nucleocapsid protein.
17. Zaki, A.M., Van Boheemen, S., Bestebroer, T.M., Osterhaus, A.D.M. and Fouchier, R.A.M. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia.
18. Zhong, N.S., Zheng, B.J., Li, Y.M., Poon, L.L.M., Xie, Z.H., Chan, K.H., Li, P.H., Tan, S.Y., Chang, Q. and Xie, J.P. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China.
19. Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B. and Huang, C.L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin.