
International Journal of Agriculture, Environment and Biotechnology
Citation: IJAEB: 13(4): 511-515, December 2020
DOI: 10.30954/0974-1712.04.2020.16

MICROBIOLOGY
Protein Profiling of Oreochromis spp. Epidermal Mucus Subsequent to Challenge of Common Freshwater Bacteria
ABSTRACT
Freshwater bacteria are the emerging pathogens that cause severe systemic disease in fish worldwide. Fish epidermal mucus contains innate immune components that provide the primary defense against different pathogenic microbes. The current experiment was designed to profile molecular changes of red hybrid tilapia mucus after subsequent challenge to common freshwater bacteria. Thus, to profile the epidermal mucus, 30 red hybrid tilapia Oreochromis spp. each of 150g was infected with Streptococcus agalactiae, Staphylococcus aureus, and Aeromonas hydrophila. 10 fish were infected for each bacterium, and 10 fish acted as control. Every 1, 2, 4, and 8 hours, fish body mucus was collected in order to profile and explore molecular changes after subsequent challenge towards the causative agent. Sodium Dodecyl Sulfate Polyacrylamide Gel-Electrophoresis (SDS PAGE) was used to allow the fish body mucus protein separation by mass. As a result, common protein, 14 kDa was found in all of the isolated mucus challenged. Meanwhile, a protein with a size 49 kDa, 81kDa, and 101 kDa was found as a prominent protein of Streptococcus agalactiae. The prominent protein after challenge with Staphylococcus aureus is 20 kDa, 30 kDa, 35 kDa, and 63 kDa. In contrast, the most prominent protein after challenge with Aeromonas hydrophila is 35 kDa, 40 kDa, 60 kDa. Protein profiling of mucus after 4 hours being challenged shows the best variations from the region 14–101 kDa. All of this finding is important towards better treatment and prevention of disease occurrence in Tilapia aquaculture.
Highlights
 Common protein, 14 kDa was found in all of the isolated mucus challenged.
Common protein, 14 kDa was found in all of the isolated mucus challenged.
 Protein profiling of mucus after 4 hours being challenged shows the best variations from the region 14-101 kDa.
Protein profiling of mucus after 4 hours being challenged shows the best variations from the region 14-101 kDa.
Keywords: Freshwater, Protein profiling, Streptococcus agalactiae, Tilapia
Red hybrid tilapias (Oreochromis spp.) are mainly freshwater fish inhabiting shallow streams, rivers, lakes and less commonly found living in brackish water. Oreochromis spp. was first introduced into Malaysia in the mid-1980s and was initially contemplated to be hardy and resistant to diseases. Tilapia toleratesadverse water quality, and other stressors compare to other most commercial aquaculture species.
Streptococcus agalactiae is a major piscine pathogen that is the cause of serious economic losses in many species of freshwater and estuarine fish worldwide. Streptococcus agalactiae infection of these fish was discovered in 1997 and has now been reported in many tilapia rearing cages in Pedu, Kenyir, and Pergau Lakes, which cause mortality between 60% and 70% of these populations (Siti-Zahrah et al. 2005). The occurrence of streptococcosis outbreak increases when tilapia are stressed due to non-optimal water temperature, low dissolved oxygen, high nitrite levels, and high culture densities (Shoemaker et al. 2000).
How to cite this article: Mohd, F., Nur-Nazifah, M., Ridzuan, S., Norazlan, W., Abdullah, A. and Ramly, R. 2020. Protein Profiling of Oreochromis spp. Epidermal Mucus Subsequent to Challenge of Common Freshwater Bacteria. IJAEB, 13(4): 511–515.
Source of Support: None; Conflict of Interest: None 
Another bacterial disease that has also significantly impacted production at some farms is the disease caused by the bacteria Aeromonas hydrophila and Staphylococcus aureus. Aeromonas hydrophila is a worldwide, free-living, Gram-negative bacterium prevalent in fresh and brackish water systems. Aeromonas hydrophila is also associated with disease in carp, channel catfish, tilapia, and produces stress-related diseases in salmonids with few symptoms such as ulcerations and exophthalmia (Rahman et al. 1997).
Fish live in a microbe-rich environment and are in intimate contact with their territory. They are vulnerable to the invading pathogenic or opportunistic microorganism. Fish body mucus is the first line of defense against infections and it discriminates between pathogenic and commensal bacterial strains (Jurado et al. 2015). Mucus composition varies among fish species and serves as a wall influenced by endogenous and exogenous factors (Elavarasi et al. 2013). The mucus composition is very complex and includes numerous antibacterial factors secreted by fish's skin cells, such as immunoglobulins, agglutinins, lectins, and lysozymes. Mucus secreted by animal's goblet cells is also essential for respiration, excretion, reproduction, and osmotic regulation. The main structural proteins of mucus are high molecular mass (~106 kDa) glycoproteins called mucins (Tabak 1995). Mucins are strongly adhesive and form a matrix in which a diverse range of antimicrobial molecules can be found.
There are also numerous studies on innate immune factors in fish epidermal mucus, including the role of proteases and antibacterial agents (De Veer et al. 2007; Subramanian et al. 2007). The present study was carried out to study the profile of protein isolated from tilapia body mucus after been challenged with Streptococcus agalactiae, Staphylococcus aureus, and Aeromonas hydrophila.
MATERIALS AND METHODS
Fish preparation
Fish infection
40 red tilapia Oreochromis spp. each of average weight 150g were divided intofour identical tanks (10 fish per tank). Fish were randomly tested and screened to ensure that they were disease and pathogen-free. Three tanks were challenged with different pathogen Streptococcus agalactiae (7.7 * 10 CFU/ml), Staphylococcus aureus(1.295 * 10 CFU/ml) and Aeromonas hydrophila (9.25 * 10 CFU/ml). Every 1, 2, 4 and 8 hours, fish body mucus was collected. Mucus was carefully scraped from the fish body surface using a sterile plastic spatula. The skin mucus was harvested and immediately frozen to prevent any external bacterial contamination. Mucus sample was stored at -20°C until further use.
SDS-PAGE
Concentrated stored fish body mucous was used for sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) to compare their protein profiling. The SDS-PAGE protocol was performed based on the method of Laemmli (1970). Fish body mucous sample was mixed 1:1 with SDS-PAGE sample buffer with 2-fi-mercaptoethanol, and samples were separated by electrophoresis at 100 V in a Mini electrophoresis cell (Bio-Rad). Following electrophoresis, the gels were stained for 20 minutes in a Commassiebrilliant blue solution and destained in a solution containing 40% methanol and 7% acetic acid. Gels were scanned for image processing.
RESULTS AND DISCUSSION
Mucus represents an important biological interface between Orechromis spp. and their aqueous environment. Fish mucus plays a dual role as a component of the innate immune mechanism. First, by being constantly secreted and cast off regularly, it prevents pathogen adherence (Subramanian et al. 2007). Second, it also essentials as a repository of numerous innate immune factors such as lysozyme, immunoglobulins, complement proteins, lectins, C-reactive protein, proteolytic enzymes, and various other antibacterial proteins and peptides (Shepherd 1994; Cole et al. 1997). Accordingly, it was decided to study the proteins in this fish mucus to determine if changes occurred in these proteins during bacterial infection. SDS-PAGE is a technique frequently used in genetics, biochemistry, and molecular biology to separate proteins according to their electrophoretic mobility. Protein electrophoresis has been of great value for delineation of numerous bacterial classification and identification (Vauterin et al. 1990). It is also widely acknowledged that the electrophoretic separation of cellular proteins is a sensitive technique that mainly provides information on the similarity of the strains at and below the species level.
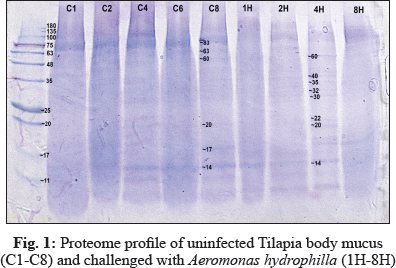
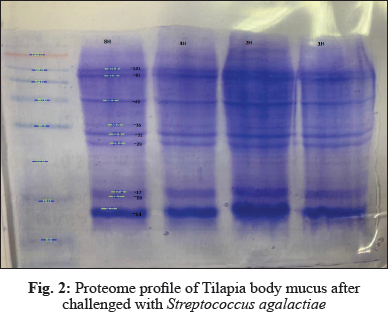
In this study, electrophoresis of fish mucus protein is used in the identification of the three different bacteria species. Each bacteria species which that has been challenged into Oreochromis spp. had characteristically distinctive body mucus protein band patterns. The clear distinct bands as 14 kDa, 20 kDa, 22 kDa, 30kDa, 32 kDa, 35 kDa, 40 kDa, and 60 kDa in mucus sample challenged by Aeromonas hydrophila (Fig. 1); 14 kDa, 16 kDa, 17kDa, 29 kDa, 32 kDa, 36 kDa, 49 kDa, 81 kDa, and 101 kDa in mucus sample challenged by Streptococcus agalactiae (Fig. 2); 14 kDa, 17 kDa, 18 kDa, 20 kDa, 30 kDa, 35 kDa, 48 kDa and 63 kDain mucus sample challenged by Staphylococcus Aureus (Fig. 3).
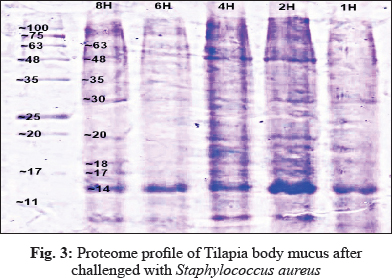
The electrophoretic analysis of fish body mucus extract revealed the presence of proteins at various molecular masses resolved by Bis-Tris 4-12% NuPAGE and Commasiebrilliant blue stained. Gels display good variations of mucus protein profile after 4 hours of challenge with all three bacteria. Based on Fig. 1, the most prominent protein after challenge with Aeromonas hydrophila is 35 kDa, 40 kDa, 60 kDa. Meanwhile, a protein with size 49 kDa, 81 kDa, and 101 kDa was found as a prominent protein of Streptococcus agalactiae (Fig. 2). The prominent protein after challenge with Staphylococcus aureus is 20 kDa, 30 kDa, 35 kDa, and 63 kDa (Fig. 3). As a result, common protein, 14 kDa was found in all of the isolated mucus challenged. Fish skin mucus is anessential component of the piscine immune system. Sarmasik (2002) suggested that the epidermal mucus acts as a first line of defense against pathogens. Fish mucus, secreted by goblet cells, plays an imperative role in nonspecific immunity by acting as a chemical or physical frontier against infectious microorganisms (Dash et al. 2018). Numerous inflammatory and antibacterial factors like immunoglobulin, proteases and lectins have been discovered in fish mucus (Rajan et al. 2013).
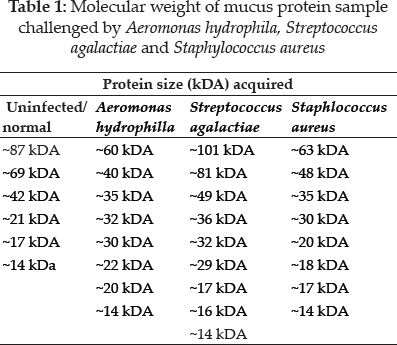
The most prominent proteins after challenge with Aeromonas hydrophila are 35 kDa, 40 kDa, 60 kDa. Meanwhile, a protein with a size 49 kDa, 81 kDa, and 101 kDa was found as a prominent protein of Streptococcus agalactiae. The prominent protein after challenge with Staphylococcus aureus is 20 kDa, 30 kDa, 35 kDa, and 63 kDa. Differences of prominent protein proving that the antimicrobial activity of skin mucus differs after challenged with three different bacteria. Post-infection, red hybrid tilapia epidermal mucus cells continuously secrete gel that forms a layer of adherent that acts as a barrier (Koshio 2016). Epidermal fish mucus able to inhibit the growth of bacteria, and therefore, the mucus may have a potential source of novel antimicrobial peptides in it (Subramanian 2007). Antimicrobial peptides (AMPs), which are the host defense peptide, are an evolutionarily conserved component of innate immunity that can function without either high specificity or memory. The peptides are synthesized at low metabolic costs, capable of mass storage, and readily available after infection. Such molecules are well suited for interacting with bacterial membranes having negatively charged and hydrophilic head groups and hydrophobic cores (Dash et al. 2018). Based on protein profiling, prominent protein indicates which antibacterial peptides interact strongly and permeate the phospholipid membranes of each different bacteria. These prominent proteins,which are unique and differ from each other, may be suitable as molecular markers for the identification and determination of various polypeptide bands of different bacteria. Thus, prominent proteins that were expressed after been challenged by different bacteria might contain antigenic properties towards the pathogen.
In uninfected red hybrid tilapia and all of the isolated mucus challenged, 14 kDa was identified as a common protein. The common protein existed as the main component protein of red hybrid tilapia mucus under normal conditions. Its composition and rheological properties are important for mucus to maintain its function,including respiration, osmoregulation, communication, locomotion, and disease resistance (Lai et al. 2009). During stress conditions, the amount of skin mucus secretion is increased, and disrupts microbial homeostasis in fish that increases the susceptibility of fish to infections (Boutin et al. 2013). This explains the changes of prominent protein after pathogen infection instead of the common protein. The composition of skin mucus also differs among fish species and production is influenced by various exogenous and endogenous factors (Esteban 2012). It can be concluded that 14 k Daprotein is the common protein in Oreochromis spp. mucus.
Protein profiling of mucus after 4 hour being challenged shows the best variations from the region 14–101 kDa. The appearance of protein perhaps due to completeness of bacteriolytic activity in fish skin mucus and other tissues contributes to its host defence mechanism against bacterial infection. Mucus is continuously being produced and removed from the integumental surface, thus physically trapping and averting bacteria from attaching the fish epithelium and invades the fish's tissue. Previous studies proved that a variety of antimicrobial proteins from fish mucus was potentially involved in the protective function against invading pathogens (Subramanian et al. 2007). Thus, mucus plays a vital role in maintaining fish health, providing a physical and biochemical barrier between the animal and the environment.
CONCLUSION
Analysis of one dimensional SDS-PAGE of Orechromis spp. body mucus challenged by Streptococcus agalactiae, Staphylococcus aureus and Aeromonas hydrophila clarifying the differences of protein profiling as each species bacteria characteristically displays distinctive protein band patterns. This study discovered 14kDa as the common protein in red hybrid tilapia mucus. Further work needs to be done to study the characterization and immunogenicity of the above antigenic proteins of mucus challenged by Streptococcus agalactiae, Staphylococcus aureus, and Aeromonas hydrophila.
ACKNOWLEDGEMENTS
The author thanks Asst. Prof. Dr. Nur Nazifah Mansor for providing experimental supports in the Department of Marine Science, Kulliyah of Science, International Islamic University Malaysia, Bandar Indera Mahkota, Jalan Sultan Ahmad Shah, Kuantan, 25200 Pahang, Malaysia. The author also thanks the National Fish Health Research Centre (NaFish), Jalan Batu Maung, 11960 Batu Maung, Pulau Pinang, Malaysia for equipment and financial support.
REFERENCES
Boutin, S., Bernatchez, L., Audet, C. and Derôme, N. 2013. Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PLoS One, 8(12): e84772.
Cole, A.M., Weis, P. and Diamond, G. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biochem., 272: 12008-12013.
Dash, S., Das, S.K., Samal, J. and Thatoi, H.N. 2018. Epidermal mucus, a major determinant in fish health: a review. Iran J. Vet. Res., 19: 72–81.
De Veer, M.J., Kemp, J.M. and Meeusen, E.N.T. 2007. The innate host defence against nematode parasites. Parasite Immunol., 29: 1–9.
Elavarasi, K., Ranjini, S., Rajagopal, T., Rameshkumar, G. and Ponmanickam, P. 2013. Bactericidal proteins of skin mucus and skin extracts from fresh water fishes, Clarias batrachus and Tilapia mossambicus. Thai J. Pharm. Sci., 37: 194–200.
Esteban, M.A. 2012. An overview of the immunological defenses in fish skin. ISRN Immunol., pp. 1–29.
Jurado, J., Fuentes-Almagro, C.A., Guardiola, F.A., Cuesta, A., Esteban, M.A. and Prieto-Alamo, M.J. 2015. Proteomic profile of the skin mucus of farmed gilthead seabream (Sparus aurata). J. Proteomics, 120: 21–34.
Koshio, S. 2016. Immunotherapies targeting fish mucosal immunity-Current knowledge and future perspectives. Frontiers in Immunology, 6: 643. spp. Epidermal Mucus Subsequent to Challenge…
Lai, S.K., Wang, Y.Y., Wirtz, D. and Hanes, J. 2009. Micro- and macrorheology of mucus. Adv. Drug Deliv. Rev., 61: 86–100.
Rahman, M.H., Kusuda, R. and Kawai, K. 1997. Virulance of starved Aeromonas hydrophila to cyprinid fish. Fish Pathology, 32: 163–168.
Rajan, B., Lokesh, J., Kiron, V. and Brinchmann, M.F. 2013. Differentially expressed proteins in the skin mucus of Atlantic cod (Gadus morhua) upon natural infection with Vibrio anguillarum. BMC Vet. Res., 9(1): 103.
Rakshit, A., Singh, H.B., Singh, A.K. and Singh, U.S. 2020. New frontiers in Stress Management for Durable Agriculture. Springer Nature, Singapore.
Sarmasik, A. 2002. Antimicrobial peptides: a potential therapeutic alternative for the treatment of fish diseases. Turkish Journal of Biology, 26(4): 201–207.
Shapiro, A.L., Vineula, E. and Maizel, J.V., Jr. 1967. Biochem. Biophys. Res. Commun., 28(8): 15–820.
Shepherd, K.L. 1994. Functions for fish mucus. Rev. Fish Biol. Fish, 4: 401-429.
Shoemaker C.A., Evans J.J. and Klesius P.H. 2000. Density and dose: factors affecting mortality of Streptococcus iniae infected tilapia (Oreochromis niloticus). Aquaculture, 188: 229-235.
Siti-Zahrah, A., Padilah, B., Azila, A., Rimatulhana, R. and Shahidan, H. 2005. Multiple streptococcal species infection in cage-cultured red tilapia, but showing similar clinical signs. In: Proceedings of the Sixth Symposium on Diseases in Asian Aquaculture. MG Bondad-Reantaso, CV Mohan, MCrumlish, RP Subasinghe, Eds., Colombo, Sri Lanka, pp. 332–339.
Subramanian, S., MacKinnon, S.L. and Ross, N.W. 2007. A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp. Biochem. Physiol. Biochem. Mol. Biol., 148: 256–263.
Tabak, L.A. 1995. In defense of the oral cavity: structure biosynthesis and function of salivary mucins. Annu. Rev. Physiol., 57: 547–564.
Vauterin, L., Swings, J., Kersters, K., Gillis, M., Mew, T.W., Schroth, M.N. … and Bradbury, J.F. 1990. Towards an improved taxonomy of Xanthomonas. Int. J. Systematic and Evolutionary Microb., 40(3): 312–316.